Electro Dynamics of Micro-Structures
The interest to optical signal detection of nanoscale objects is growing rapidly. Theoretical modeling of optical scattering by nano-particles is complementary to the development of experimental techniques. We conduct numerical computations of scattering by objects which are both comparable to and much smaller than the wavelength of light. Currently, theoretical analysis of both single protein and retina scattering is underway. The employed techniques can be extended to the analysis of many other types of structures.
The retina contains scattering elements that are comparable in size to the wavelength of light. It is comprised of photoreceptor cells, which act as dielectric wave-guides. This natural microstructure has many interesting properties. Numerical modeling of the retina reveals complex interactions of different optical modes. This interaction may influence the visual response of an organism [Figure 1]. Furthermore, analysis of scattering in polarization sensitive retina (such as retina of birds and fish) may contribute to progress in the development of optical sensors. The retina is modeled using the Finite-Difference Time-Domain (FDTD) numerical algorithm. This research is a result of collaboration with Dr. Misha Vorobyev from the Center for Vision, Touch and Hearing of the University of Queensland and Dr. Andrey Zvyagin from Macquarie University.
Figure 1. Longitudinal cross-section through human foveal cone photoreceptor model. Time-averaged optical intensity is shown for different wavelengths, with illumination from the left. Lighter regions indicate higher intensity (colour axis is 0 to 15). Oscillations are the result of mode interaction.
Scattering by a single protein molecule has been calculated by our group using the Discrete Dipole Approximation (DDA). Our partners on this project are Dr Timo Nieminen and Mr Vincent Loke from the University of Queensland. This work is motivated by the potential of optical detection of the folding state of a single protein. Although the scattering cross-section of a single protein is incredibly small, the signal contrast available between folded and unfolded states has been found to be significant [Figure 2]. Local effects within the protein environment are also of interest. The polarisability of a protein region varies depending on its location and chemical composition, hence influencing its interaction with an incident field. This is expected to affect a number the of nanoscopic optical characteristics, such as the Förster distance in FRET between two chromophors embedded in protein made environments.
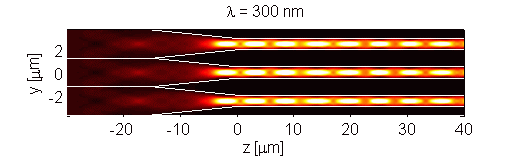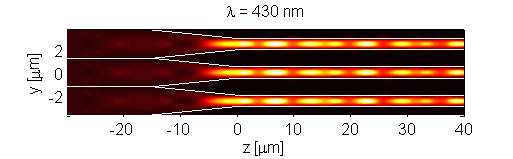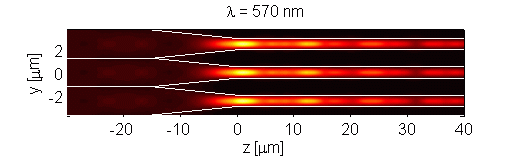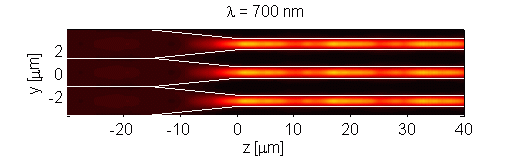
Figure 2. Integral scattering cross-section of folded and unfolded proteins as a function of molecular weight. Scattering cross-section was determined via DDA for standard molecular weight markers (PDB ID shown). Scattering cross-section is proportional to the square of molecular weight, with proportionality constant being smaller for unfolded